Free Courses Sale ends Soon, Get It Now


Free Courses Sale ends Soon, Get It Now



Disclaimer: Copyright infringement not intended.
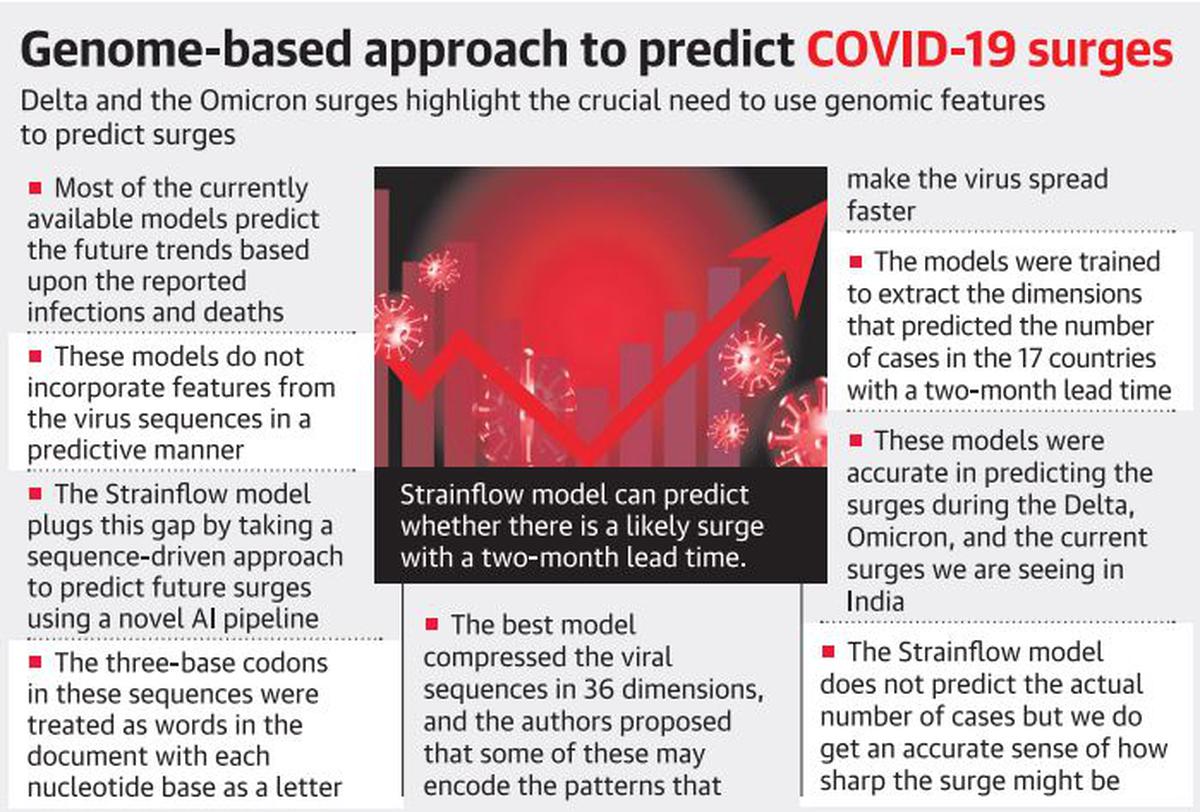
Introduction
Strainflow Model
© 2024 iasgyan. All right reserved